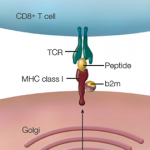
The group of 440 consortium scientists performed 24 different types of experiments to examine and identify regions of transcription, transcription factor association, chromatin structure, and histone modification in various cell types. Most of the experiments were performed with cell lines such as HeLa, GM12878, K562 and HUVEC; a major (and very necessary) goal to be accomplished is to examine primary cells from both people with specific diseases and healthy individuals. Few experiments have been performed that specifically examined cells of the immune system (subsets of T cells, B cells) or islets, liver, heart, intestine, lung, and kidney cells, but we can expect these blanks to be filled in within the next few years.
The data produced by the ENCODE consortium, which includes 5 trillion bytes of raw data representing more than 1,640 genome-wide data sets from 147 cell types, is still far from complete. All ENCODE data are freely available for download and analysis at the ENCODE data coordination center at the University of California, Santa Cruz (genome-preview.ucsc.edu/encode).
So—how to navigate the massive amount of data? The published manuscripts are all freely available in the ENCODE explorer (nature.com/encode) or with the very nifty ENCODE iPad App. In addition to the individual papers, the findings are organized into “threads” so that all of the relevant insights from all the publications are contained in one document. Only time will tell whether the massive amount of data obtained in the ENCODE project will translate into improvements in the diagnosis and treatment of human disease.
Reprinted with permission from Am J Transplant. 2013; 13:245.
Dr. Krams is associate professor of surgery at Stanford School of Medicine in Stanford, Calif. Dr. Bromberg is professor of surgery and microbiology and immunology, and is the chief of the division of transplantation at the University of Maryland Medical Center in Baltimore.