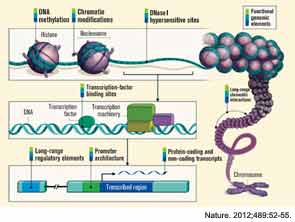
The ENCODE project provides information beyond that contained within the DNA sequence—it describes the functional genomic elements. The project contains data about the degree of DNA methylation and chemical modifications to histones that can influence the rate of transcription of DNA into RNA molecules (histones are the proteins around which DNA is wound to form chromatin). ENCODE also examines long-range chromatin interactions, such as looping, that alter the relative proximities of different chromosomal regions in three dimensions and also affect transcription. The project describes the binding activity of transcription-factor proteins and the location and sequence of gene-regulatory DNA elements, which include the promoter region upstream of the point at which transcription of an RNA molecule begins, and distant, long-range regulatory elements. One section of the project mapped DNase I hypersensitive sites, indicating specific sequences at which the binding of transcription factors and transcription-machinery proteins has caused nucleosome displacement.
Advances in genomics and bioinformatics have provided critical new insights into our understanding of the human genome—especially in identifying protein-coding genes—yet questions remain, including the functionality of the large stretches of noncoding DNA. Recently scientists in 32 labs around the world, as part of the Encyclopedia of DNA Elements (ENCODE) project, have reported in a staggering 30 papers in the journals Nature, Genome Biology, and Genome Research that the widely held belief that large segments of the human genome consists of “junk” DNA is incorrect. Findings from the ENCODE project reveal that 80% of the noncoding DNA does indeed have a biochemical function in at least one cell type. Contained within the noncoding roadmap of the genome are promoters, enhancers, and regions that encode RNA transcripts that have regulatory functions but are not translated into proteins (see Figure 1). The implications of these findings are protean in helping us to understand human variation and how changes in the genome result in disease.
Genome-wide association studies (GWAS) have indicated that a large proportion of the single-nucleotide polymorphisms (SNPs) correlated with disease phenotype are actually located in introns or within noncoding sequences. When the ENCODE consortium examined more than 4,500 SNP phenotype associations over a range of human diseases, they determined that 12% of these SNPs overlap regions that contain transcription factors, and 34% overlap DNase I hypersensitive sites, which are transcriptionally active DNA. For example, the SNP rs11742570, which has been shown to be strongly associated with Crohn’s disease, overlaps a GATA2 transcription-factor–binding signal. These findings tell us that many disease-associated changes are not actually in the genes themselves, but rather in the regions that regulate the genes. Clearly, interpretation of GWAS results must consider both the coding and noncoding regions of the genome.
The group of 440 consortium scientists performed 24 different types of experiments to examine and identify regions of transcription, transcription factor association, chromatin structure, and histone modification in various cell types. Most of the experiments were performed with cell lines such as HeLa, GM12878, K562 and HUVEC; a major (and very necessary) goal to be accomplished is to examine primary cells from both people with specific diseases and healthy individuals. Few experiments have been performed that specifically examined cells of the immune system (subsets of T cells, B cells) or islets, liver, heart, intestine, lung, and kidney cells, but we can expect these blanks to be filled in within the next few years.
The data produced by the ENCODE consortium, which includes 5 trillion bytes of raw data representing more than 1,640 genome-wide data sets from 147 cell types, is still far from complete. All ENCODE data are freely available for download and analysis at the ENCODE data coordination center at the University of California, Santa Cruz (genome-preview.ucsc.edu/encode).
So—how to navigate the massive amount of data? The published manuscripts are all freely available in the ENCODE explorer (nature.com/encode) or with the very nifty ENCODE iPad App. In addition to the individual papers, the findings are organized into “threads” so that all of the relevant insights from all the publications are contained in one document. Only time will tell whether the massive amount of data obtained in the ENCODE project will translate into improvements in the diagnosis and treatment of human disease.
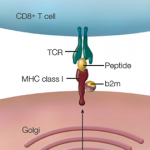
Reprinted with permission from Am J Transplant. 2013; 13:245.
Dr. Krams is associate professor of surgery at Stanford School of Medicine in Stanford, Calif. Dr. Bromberg is professor of surgery and microbiology and immunology, and is the chief of the division of transplantation at the University of Maryland Medical Center in Baltimore.