SAN DIEGO—The inflammatory myopathies are a heterogeneous group of conditions that, although discovered decades ago, continue to challenge rheumatologists in terms of their myriad clinical presentations. In Flexing Strong Science on Weak Muscles: Genetics, Genomics and Autoantibodies in Myositis, two speakers provided exceptionally helpful insights into these conditions, using novel tools for analysis.
6 Types
The first speaker was Andrew Mammen, MD, PhD, senior investigator and chief, Muscle Disease Section, National Institute of Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, Md. Dr. Mammen began by describing the six identifiable types of myositis.
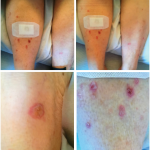
Inclusion body myositis (IBM) is known for its unique features, such as weakness in the distal finger flexors and a muscle biopsy that may show inflammatory cells surrounding and invading non-necrotic muscle fibers, rimmed vacuoles, protein aggregates and congophilic inclusions. However, Dr. Mammen noted that these classic findings on muscle biopsy may not be present in a sizable number of patients and, thus, the absence of these findings should not rule out the diagnosis.
Dermatomyositis (DM) is classically associated with perifascicular damage on muscle biopsy, and there now exist many identifiable myositis-specific autoantibodies that can be identified, each with its own characteristic clinical phenotype.
The muscle biopsies in patients with anti-synthetase syndrome (AS) are hard to distinguish from those of patients with DM, but a key feature of patients with this condition may include significant interstitial lung disease and particular cutaneous findings, like mechanic’s hands.
Muscle biopsies in patients with immune mediated necrotizing myopathies (IMNM) will show myofiber necrosis, and the autoantibodies that track with this condition are those directed against signal recognition particle (SRP) and hydroxymethylglutaryl CoA reductase (HMGCR).
There are overlap forms of myositis, in which patients may have features of conditions, such as systemic lupus erythematosus, systemic sclerosis and rheumatoid arthritis, and muscle biopsies in these patients are variable. Finally, it has been noted that certain patients exposed to immune checkpoint inhibitors (ICI) for treatment of malignancies, such as lung cancer or melanoma, may develop a form of myositis as an adverse effect of these medications.
Research Questions
Dr. Mammen went on to describe the work of his research team in easy-to-understand terms: chunks of muscle biopsy tissue are collected (from patients both with and without myositis), RNA is extracted from this tissue, and a genetic library is prepared. RNA sequencing is performed and produces an expression level of every identifiable gene. Dr. Mammen asked several questions in his lecture and was able to provide answers based on his research findings.
First: do different types of myositis have different interferon (IFN) gene signatures? The answer to this question appears to be yes. In a paper from Pinal-Fernandez and colleagues, the expression of IFN1-inducible genes was high in patients with DM, moderate in patients with AS, and low in patients with IMNM. With regards to IFN2-inducible genes, these are high in patients with DM, IBM, and AS but appear to be low in those with IMNM.1 These IFN gene signatures may prove to be helpful both in understanding the pathophysiology of disease and in identifying treatment targets for future therapeutics.
The second question that Dr. Mammen asked was: Can transcriptomic data be used to make a diagnosis? For this, he pointed to additional work by Pinal-Fernandez and colleagues that shows how transcriptomic analysis may be better than muscle biopsy-based histopathology in making a diagnosis in patients with myositis. In one study, RNA sequencing was applied to muscle biopsies in 119 patients with myositis (either IBM or defined myositis specific autoantibodies) and 20 controls. Machine learning algorithms were trained using transciptomic data, and specific recursive feature elimination was employed to identify the genes most useful for classifying muscle biopsies into each type and MSA-defined subtype of myositis. Ultimately, the machine learning algorithm classified the muscle biopsies with more than 90% accuracy.2
Research studies such as this indicate that the marriage between transcriptomic analysis and machine learning may prove to be fruitful in correctly diagnosing many patients with specific forms of myositis in the future.
The third question that Dr. Mammen sought to answer was: is ICI-myositis one entity? Again, the work of Pinal-Fernandez and colleagues has shown that there are actually three types of ICI-myositis, and these are ICI-DM, ICI-MYO1 and ICI-MYO2. While the interleukin-6 (IL-6) pathway was overexpressed in all three of these groups, only ICI-MYO1 patients developed myocarditis. These patients also had highly inflammatory features on muscle biopsy. In contrast, patients with ICI-MYO2 had mostly necrotizing pathology and low levels of muscle inflammation.3
Dr. Mammen explained that these findings help clinicians to better understand ICI-myositis as a whole, which is still a relatively newly described entity, but one that will be increasingly important as more and more ICI therapies are applied to patients with cancer.
3 New Genome-Wide Associations Identified
The second speaker in the session was Christopher Amos, PhD, professor and chief of medicine, epidemiology and population science, Baylor College of Medicine, Houston, Texas, who discussed additional novel findings regarding genetic analysis in patients with myositis. In a significant paper from this past year, Dr. Amos and colleagues analyzed more than 2,500 samples from white patients with idiopathic inflammatory myopathies (IIM) collected through the Myositis Genetics Consortium (MYOGEN), as well as more than 10,000 ethnically matched control samples. Dr. Amos and colleagues then imputed over 1.6 million variants from the ImmunoChip array using the Haplotype Reference Consortium panel and conducted an association analysis.
Through the use of imputation to identify and fine-map genetic associations in IIM, the authors were able to identify three new genome-wide associations across all patients: STAT4, SDK2, and LINC00924. They also identified a novel genome-wide association with NAB1 in the subgroup of patients with polymyositis. Finally, the HLA region was found to be the most significant genetic risk factor for the entire IIM group, and the most significant risk in this region was seen in the group with antibodies to Jo-1. This is important since the study represents the largest genetic association study to date investigating non-HLA genes in patients with anti-Jo-1 autoantibodies.4
Dr. Amos was able to describe research techniques that are novel and interesting, and clearly the future of investigation on myositis will continue to employ these strategies.
In Sum
The entirety of the session was informative and exciting, and the audience was provided both with clinical pearls and food-for-thought regarding future research directions. The effects of myositis may be to cause weakness, but the strengths of this session can be found in the light that is now shed on understanding myositis in all its many forms.
 Jason Liebowitz, MD, is an assistant professor of medicine in the Division of Rheumatology at Columbia University Vagelos College of Physicians and Surgeons, New York.
Jason Liebowitz, MD, is an assistant professor of medicine in the Division of Rheumatology at Columbia University Vagelos College of Physicians and Surgeons, New York.
References
- Pinal-Fernandez I, Casal-Dominguez M, Derfoul A, et al. Identification of distinctive interferon gene signatures in different types of myositis. Neurology. 2019;93(12):e1193–e1204.
- Pinal-Fernandez I, Casal-Dominguez M, Derfoul A, et al. Machine learning algorithms reveal unique gene expression profiles in muscle biopsies from patients with different types of myositis. Ann Rheum Dis. 2020;79(9):1234–1242.
- Pinal-Fernandez I, Quintana A, Milisenda JC, et al. Transcriptomic profiling reveals distinct subsets of immune checkpoint inhibitor induced myositis. Ann Rheum Dis. 2023;82(6):829–836.
- Rothwell S, Amos CI, Miller FW, et al. Identification of novel associations and localization of signals in idiopathic inflammatory myopathies using genome-wide imputation. Arthritis Rheumatol. 2023;75(6):1021–1027.