
During the past five years, an explosion of information has emerged from genetic association studies of systemic lupus erythematosus (SLE). The sheer number of genes identified through this work, as well as the specific genes that have been identified as SLE risk factors, demonstrate the surprising degree of success of genome-wide association and related studies to reveal the genetic underpinnings of this complex disorder. Further, the remarkable degree of clustering of recently identified variants into specific biologic pathways has, to a greater extent than anticipated, provided insights into the etiologic processes that contribute to SLE. Also surprising has been the extent to which recently identified genetic variants appear to be relevant to multiple autoimmune diseases, highlighting the close etiologic relationships among this broad and clinically diverse group of human disorders. Given the rapid pace of gene discovery in SLE, it is helpful to review the relevant concepts and findings of this research, including what they reveal about this complex disorder. In addition, I also will briefly describe in this article the direction of future research in this area.
A Genetically Complex Disorder
Although the prevalence of SLE is relatively low, recent genetic discoveries highlight the close etiologic relationships among the large and diverse group of human autoimmune disorders. These findings are consistent with the well-known clustering of these diseases within families and individuals. The genetic complexity of SLE means that the disease results from the action of many predisposing genes as well as environmental and other nongenetic factors. As a consequence of this complexity, SLE does not demonstrate classical Mendelian modes of familial inheritance, and there is a lack of correspondence between the genotype that one inherits and disease status. Complex genetic disorders also are characterized by heterogeneity of genetic associations, according to ethnic and clinical differences, for example; this heterogeneity has been strongly reinforced by recent genetic studies of SLE.
In spite of these complexities, there is clear evidence of familial clustering for SLE, supporting an important role for genetics. For example, siblings of SLE patients are approximately 30 times more likely to develop SLE compared with individuals without an affected sibling. This ratio is referred to as ls and is notably higher in SLE compared with many other autoimmune diseases. For example, the ls for psoriasis and rheumatoid arthritis is in the range of 5–10. Although the relatively larger ls for SLE should theoretically translate into a greater ease of identifying SLE genes, it is important to remember that this parameter reflects the action of all genetic risk factors. Thus, the ease with which SLE genes can be identified will vary greatly depending upon whether the increased risk of disease in families is due to the action of 30 genes versus 300 or even 3,000 genes.
Genome-Wide Association Studies
The pace of gene discovery in SLE has increased exponentially during the past five years due primarily to the increased feasibility of performing very large genome-wide association studies (GWAS) using hundreds of thousands of single-nucleotide polymorphism (SNP) markers. (See Definitions (at right) for a brief definition of SNP and other technical terms used here.) The first GWAS of a complex human disease was completed in 2006, and since then, application of this methodology to SLE has resulted in the discovery of over 20 firmly established risk loci. This rapidity of recent progress is in contrast to the previous slow pace of gene discovery that began in the 1970s with the discovery of HLA gene associations with SLE and was followed by additional discoveries that largely paralleled developments in molecular genetic technologies.1
Definitions
- Single-nucleotide polymorphism (SNP): Genetic variation in a DNA sequence that occurs when a single nucleotide is altered. SNPs may be further classified according to whether the variation occurs within an exon (exonic, or coding SNP), intron (intronic SNP), or noncoding region.
- High-throughput sequencing: High-throughput DNA sequencing technologies parallelize the sequencing process, producing thousands or millions of sequences at once, resulting in dramatically lower costs compared to standard DNA sequencing methods.
- Copy-number variation (CNV): A segment of DNA in which copy-number differences have been found by comparison of two or more genomes. The segment may range from one kilobase to several megabases in size.
- DNA methylation: The addition of a methyl group to DNA, for example, to the number 5 carbon of the cytosine pyrimidine ring. This modification can be inherited through cell division.
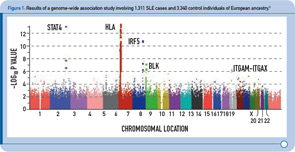
During the past two years, four GWAS of SLE cases and controls of European descent have been published, followed by two GWAS of SLE cases and controls of Asian ancestry published more recently.2–7 Figure 1 (above) summarizes the results from one of these studies and provides a graphical representation of the key features of these study designs.2 In brief, each dot in this figure (known as a Manhattan plot due to the similarity to the Manhattan skyline) corresponds to a genetic marker that, in this particular study, included ~550,000 SNPs. The dots are color coded and arranged along the x-axis according to position (with each color representing a different chromosome). The y-axis represents the significance level (–log P value) for the association of each SNP with SLE (i.e., comparison between SLE cases and controls). Given the size of this study, with more than 500,000 SNPs, the multiple testing burden is quite high and therefore the level of significance for definitive genetic associations is considered to be approximately 5 x 10–8, with results falling between –log P values of approximately 5–7.3 representing associations of borderline significance. The latter findings have been the intense focus of follow-up investigations, as described below. Thus, on the basis of this single experiment, which included about 1,300 SLE cases and approximately 3,300 control individuals of European ancestry, the HLA, STAT4, and IRF5 loci exceeded the level of significance for definitive results, and the top novel loci were BLK and ITGAM-ITGAX, which exceeded this threshold following genotyping in additional SLE cases and controls of European ancestry.

