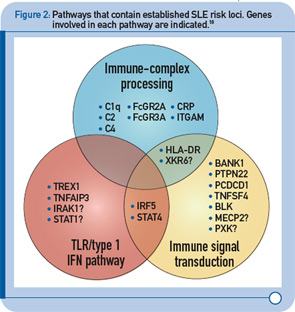
More recently, a large group of investigators from the United States and Sweden performed a follow-up study of the top loci from the aforementioned GWAS to identify additional risk loci.8 More specifically, the investigative team genotyped more than 3,000 SNPs from approximately 2,500 distinct regions that showed nominal evidence of association with SLE (P < 0.05) in an independent sample of about 2,000 SLE cases and about 4,300 controls. This replication effort identified five new SLE susceptibility loci (with p < 5 x 10–8) in TNIP1, PRDM1, JAZF1, UHRF1BP1, and IL10. Of interest, but not unexpected for a genetically complex disease, the strength of association for these loci was modest, with odds ratios ranging from 1.17 (for UHRF1BP1) to 1.27 (for TNIP1). Also as expected based on characteristics of the markers chosen for genotyping, the associated variants were relatively common, with minor allele frequencies greater than 5%. This study also identified 21 candidate loci with P ≤ 1 x 10–5, which will undoubtedly be the focus of additional studies. Lastly, these authors analyzed alleles previously associated with other autoimmune diseases and found evidence to support association with SLE for five additional loci (P < 1 x 10–3), including IFIH1, CFB, CLEC16A, IL12B, and SH2B3. These results, together with the prior GWAS and other recent studies have expanded the number of established SLE susceptibility loci to approximately 30.9 Importantly, these recent findings implicate several key immunologic pathways in SLE pathogenesis on the basis of the clustering of these genes into specific biologic pathways, including those related to immune complex processing, immune signal transduction, and the toll-like receptor (TLR) and type 1 interferon (IFN-1) pathways (see Figure 2, p. 31).10
TLR and IFN-1 Pathways
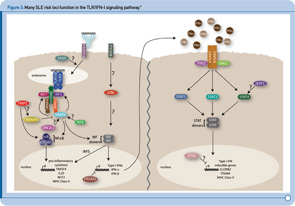
Recent genetic discoveries highlight importance of these two pathways in SLE. Perhaps most striking has been the extent to which recent discoveries in SLE highlight the importance of the TLR and IFN-1 signaling pathways.9 For example, Figure 3 (p. 31) shows key components of these interrelated pathways, many of which have now been firmly established as SLE risk loci based on recent genetic studies.
One of the key components of these pathways, TNFAIP3, nicely illustrates the extent to which some of the recently identified risk loci are clearly relevant to multiple autoimmune disorders. TNFAIP3, which was initially implicated in an early GWAS of Crohn’s disease, has now been implicated in genetic studies of at least six autoimmune disorders, including SLE. Thus, this gene may represent the best example of a true autoimmunity locus with relevance to a broad spectrum of disease, outside the HLA complex of genes. Many questions remain, however, about the mechanism by which genetic variation in the TNFAIP3 region contributes to autoimmunity. Illustrative of these challenges is the fact that many different variants, which span a broad genomic region, have been implicated with little overlap across diseases in the most strongly associated variants. Further, although some studies have implicated coding and intronic SNPs that may be functionally relevant, much work remains to establish the specific mechanisms by which these or other variants contribute to autoimmune disease processes. Nonetheless, compelling functional data exists for some of the recently identified SLE risk loci. Table 1 (p. 32) lists some of the genes for which evidence supporting a functional role of the associated variants has been reported.
Genetics and Disease Heterogeneity
In addition to the insights that findings related to TNFAIP3 and other genes have provided about fundamental autoimmune processes, recent work also highlights the fact that many recently established risk loci may predispose more specifically to certain types of disease. For example, following the identification of STAT4 as a genetic risk factor for rheumatoid arthritis and SLE, additional work demonstrated that the implicated STAT4 variant is associated more specifically with severe forms of SLE.11 For example, compared to the frequency of the risk variant in control individuals of 22.5%, the frequency of the risk variant was about 30% among individuals with disease characterized by oral ulcers and/or photosensitivity (odds ratio [OR] ~ 1.5), 35% among patients whose disease is characterized by anti-dsDNA autoantibody production (OR ~ 1.9), and 38% among patients with severe renal involvement (OR > 2.0). Similarly, work underway now suggests that most of the recently identified SLE risk variants are more strongly associated with anti-dsDNA positive disease compared with anti-dsDNA negative disease.12 Thus, an important area for future work will be to further refine genotype–phenotype associations in SLE in order to dissect, with greater etiologic relevance, this genetically, and clinically complex disease.
Current Status and Future Directions
In spite of the success of recent GWAS and other genetic studies, at least in terms of the number and specific loci identified, it has been surprising to realize that these loci, collectively, explain only a minority of the heritability of the disease. However, the same can be said about most complex diseases that have been the focus of GWAS and related studies over the past five years.13 Thus, an important question that is the focus of much current investigation in this area is what explains this “missing heritability.” There are a number of possibilities, many of which have already received some support.