As of today, more than 100 amino acid coding variants of HLA-B27 have been described. With regard to AS disease association, they fall into three categories:12
- Definitely associated with disease (including 27:02, 27:04, 27:05);
- Not associated with disease and possibly protective (including 27:06, 27:09); or
- Unknown association.
The third group contains the majority of the identified HLA-B27 variants, simply because they are so rare individually that disease association cannot be reliably determined. Sequence analysis suggests that HLA-B*27:05 is the ancestral allele and that the other alleles have evolved from this sequence.13 In the case of the susceptible HLA-B*27:05 and the non-susceptible HLA-B*27:09, the difference is only a single amino acid (amino acid 116 aspartate -> histidine).12
HLA-B27 Assays
Although HLA-B27 is a genetic marker, the HLA-B27 status of a person can be determined using antibody-based cellular assays or with nucleic acid-based techniques (see Figure 2). The original method for HLA typing, which is still being used by some histocompatibility testing laboratories, was a cytotoxicity assay with monospecific sera from HLA-sensitized individuals and cells from the test subject. The binding of polyclonal anti-HLA-B27 antibodies present in these sera to HLA-B27 proteins on the cell surface results in complement-mediated lysis, which can be read out using a cell viability stain.
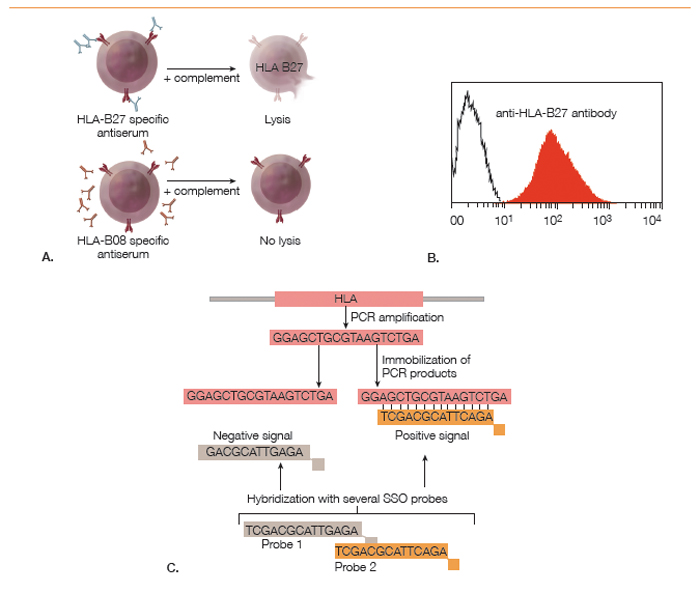
(click for larger image)
Figure 2: Methods for Determining HLA-B27 Status: A. Cytotoxicity assay using polyclonal monospecific antisera from sensitized humans; B. Flow cytometry analysis of cells stained with a fluorescently labeled monoclonal anti-HLA-B27 antibody; C. Polymerase chain reaction–sequence specific oligomerization (PCR-SSO). PCR amplification of the polymorphic exon is followed by hybridization with fluorescently labeled oligonucleotide probes to determine subtype identity.
The development of monoclonal antibodies has made HLA-B27 determination by flow cytometry feasible. In this method, peripheral blood cells from the test subject are incubated with a fluorescently labeled monoclonal antibody against HLA-B27. As MHC class I molecules are expressed on all cells, HLA-B27 positivity causes a fluorescence intensity shift of all cells relative to unstained cells or HLA-B27 negative control cells.
Antibody-based cellular techniques can determine only the serotype, which is sufficient in most clinical scenarios. Determining whether a subject is HLA-B*27:05 or HLA-B*27:09 requires a nucleic acid-based method, such as PCR-SSO (polymerase chain reaction–sequence specific oligomerization). This is a two-step method. First, a PCR reaction with common primers flanking exons 2 and 3 of the HLA-B gene is performed to amplify the polymorphic regions of the HC. In the second step, the PCR product is hybridized with a series of sequence-specific labeled oligonucleotides, which bind only in case of sequence complementarity. The hybridization pattern can then be used to infer the identity of the HLA allele. Alternatively, the nucleotide sequence of the PCR fragment is directly determined by DNA sequencing.
Worldwide Distribution of HLA-B27 Alleles
There are striking differences in the worldwide distribution of HLA-B27 and its variants (see Figure 3 and Figure 4). HLA-B27 is very rare in the southern hemisphere and also in Japan.14 It is generally much more common in the northern hemisphere, particularly in indigenous populations of the circumpolar regions. The HLA-B27 variants also show marked differences in their worldwide distribution. HLA-B*27:05 is found in central Europe, HLA-B*27:02 is common in the Mediterranean, and HLA-B*27:04 is the predominant allele in China. Among the non-associated alleles, HLA-B*27:06 is dominant in Southeast Asia, and HLA-B*27:09 is found on the Mediterranean island of Sardinia.