4) Microbial hypotheses: Multiple lines of evidence suggest the involvement of microbes in the pathogenesis of HLA-B27 dependent disease. This includes the association of HLA-B27 with reactive arthritis triggered by certain Gram-negative bacteria, the high prevalence of subclinical intestinal inflammation in AS patients and the microbiota dependence of the arthritis in HLA-B27 transgenic rats described earlier.35,36 Potential mechanisms to explain these associations include immunological cross-reactivity with microbial antigens, dysregulated defenses against intracellular bacteria in HLA-B27 expressing phagocytes or indirect effects via the intestinal microbiome.37
New Genetic Data on HLA-B27 & AS
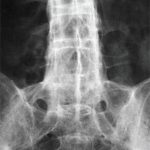
The association of MHC variants with AS was recently revisited using Immunochip data from 9,069 AS cases and 13,578 population controls.38 In this study, the sequences of the MHC genes were imputed (i.e., they were inferred based on the presence of certain local SNPs rather than determined through nucleotide-by-nucleotide sequencing). Genomic sequence imputation is possible because of linkage disequilibrium, which means that two neighboring genetic loci are likely to be inherited in a linked fashion. Therefore, by knowing the sequence (or the SNP pattern) at one locus, one also knows (with a certain probability) the sequence of the closely linked locus.
Cortes et al found, not surprisingly, that HLA-B*27:02 and HLA-B*27:05 were very strongly associated with AS. After conditioning on these two variants, they identified additional associations with non-HLA-B27 variants (i.e., HLA-B*51:01, HLA-B*47:01, HLA-B*40:02, HLA-B*13:02 and HLA-B*40:01), confirming earlier reports of additional MHC class I associations with AS. HLA-B*07:02 and HLA-B*57:01 had small protective effects. Because the study population was exclusively European, the effects of other HLA-B alleles (e.g., 27:03, 27:06, 27:09) could not be ascertained.
Cortes et al also analyzed the association of the different MHC alleles with disease at the amino acid level and found an extremely strong association with amino acid 97 (p<10-3221). This amino acid is located at the bottom of the peptide binding groove, near the so-called F pocket, where the C terminus of the peptide binds. Asparagine or threonine residues at position 97 were associated with AS, whereas serine decreased disease risk. Although the association with amino acid 97 could reflect linkage disequilibrium with the real disease-causing variants, it’s important to note that amino acid 97 is in immediate proximity to amino acid 116, a position that critically determines disease susceptibility. The AS-associated HLA-B*27:05 has aspartic acid in position 116, whereas the non-associated and otherwise sequence-identical HLA-B*27:09 allele has a histidine at position 116.
Genetic Interaction of HLA-B27 & ERAP1/2
Despite the strong association between HLA-B27 and AS, this gene has been estimated to explain only 40% of the genetic variance. Genome-wide association studies (GWAS) have revealed additional genetic associations with AS. Most relevant for this discussion, polymorphisms in ERAP1, ERAP2 and other peptidases involved in processing peptides for presentation in the MHC class I pathway were identified. ERAP1 and ERAP2 are ER-associated aminopeptidases (i.e., enzymes that trim peptides from the amino terminus prior to loading into nascent MHC molecules). ERAP1 is highly polymorphic, with many of these polymorphisms resulting in amino acid changes that affect enzyme activity.