 As the name implies, systemic lupus erythematosus (SLE) presents as a systemic disease, but it may also present in an organ-limited/dominant form. This variability makes early disease especially difficult to diagnose, and delayed diagnosis can lead to organ damage and is associated with increased mortality.
As the name implies, systemic lupus erythematosus (SLE) presents as a systemic disease, but it may also present in an organ-limited/dominant form. This variability makes early disease especially difficult to diagnose, and delayed diagnosis can lead to organ damage and is associated with increased mortality.
In many cases, diagnosis occurs because clinicians focus on the presence of a few high-yield clinical and immunologic symptoms, such as a malar rash in an individual with anti-DNA autoantibodies, that can be used in routine practice to clinically diagnose patients with SLE with high sensitivity and specificity.
Christina Adamichou, a graduate student at the University of Crete School of Medicine, Greece, and colleagues hypothesized that it may be possible to train machine learning-based tools with medical data to simulate the medical reasoning that occurs in successful diagnosis of SLE. To do this, Adamichou et al. developed and validated a new algorithm that uses classical disease features to help clinicians diagnose SLE earlier in the disease process. They describe their SLE Risk Probability Index (SLERPI) in an article published online Feb. 10 in the Annals of Rheumatic Diseases.1
“This is a novel approach to address the issue of accurately and timely diagnosis of SLE,” says Sarfaraz Hasni, MD, director of Lupus Clinical Research at the National Institute of Arthritis and Musculoskeletal and Skin Diseases at the National Institutes of Health in Bethesda, Md. “On average, SLE patients visit up to three providers, and it may take up to two years or more before they are diagnosed with SLE. This [delay] is even worse for women of color, who are disproportionality affected by more severe SLE.”
To create SLERPI, the researchers leveraged well-characterized data from the medical records of two large discovery and validation patient cohorts from two centers that maintained detailed patient registries and used structured forms for collecting clinical data. The investigators mined these data for features that could be combined with the 2019 European League Against Rheumatism (EULAR)/ACR SLE Classification Criteria to enhance diagnostic accuracy.2 Although intended to be used for research—not diagnostic—purposes, these criteria have a high degree of accuracy. The researchers identified well-defined features of SLE that were not part of the classification criteria, which they combined with the published classification criteria. They found thrombocytopenia/autoimmune hemolytic anemia (AIHA), malar rash, proteinuria, anti-nuclear antibodies (ANA), immunological disorder and combined C3 and C4 hypocomplementemia were the strongest predictors against competing rheumatological diseases. These 14 variably weighed clinical and serological features were used to train the new Lease Absolute Shrinkage and Selection Operator-logistic regression (LASSO-LR) model.
Next, the researchers used the model to calculate risk probabilities they thought corresponded with varying diagnostic certainty levels: unlikely, possible, likely and definite SLE. They used the validation cohort to assess the proportion of actual patients with SLE and control patients captured within each predicted SLE risk group. They found the calculated SLE risk probabilities correlated positively with disease severity and organ damage.
The investigators then used the discovery cohort to identify a cut-off to separate SLE from other rheumatic diseases. They chose the 50% risk probability threshold and found that at this threshold, the SLERPI demonstrated both high sensitivity (95.1%) and specificity (93.7%). SLERPI was accurate to 94.8% in the total validation cohort. When the team tested the cut-off against the control subset with undifferentiated connective tissue disease, the model specificity was 91.1%. The model also yielded very high rates of correct predictions within multiple SLE patient subgroups: for early SLE, a sensitivity of 93.8%; for lupus nephritis, a sensitivity of 97.9%; for neuropsychiatric lupus, a sensitivity of 91.8%; for hematological lupus, a sensitivity of 98.6%; for severe SLE, a sensitivity of 96.4%.
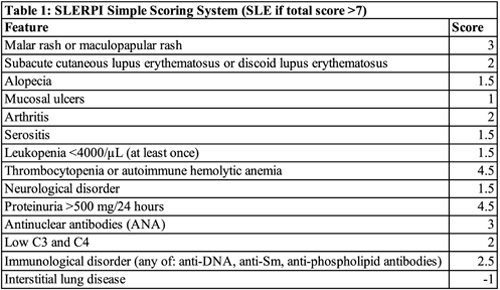
Finally, to facilitate the use of SLERPI in daily practice, the researchers converted their model to a simple scoring system. They found an operational cut-off score of 7 converted the logistic regression model into a simple scale for both clinical and serologic features of SLE (Table 1).
“Although I do not think machines can replace human/clinician assessment, I think this scoring system certainly has a role,” says Brett Smith, DO, a rheumatologist at Blount Memorial Physicians Group in Alcoa, Tenn. “If this were made available to primary care practices as a way to triage referrals to rheumatologists, it would provide a potentially reliable way to schedule referrals based on acuity or disease activity.”
Daniel J. Wallace, MD, associate director of the rheumatology fellowship program at the David Geffen School of Medicine Center, University of California, Los Angeles, disagrees. “This is an interesting study, but it really applies only to Greece,” he says. “We have a different health system, electronic medical records, and a different diversity of patients and cultures. Most of us diagnose lupus using our intuition, training and experience and don’t rely on algorithms.” He adds that not all predisposition factors for SLE will apply for all patients and he is concerned that an algorithm may lead to an inaccurate diagnosis and unnecessary treatment.
“For those practitioners whose encounters with lupus patients are infrequent and their [knowledge] from medical school rusty, the availability of a tool that promotes the inclusion of SLE in the differential diagnosis and consequently accelerates referrals to specialists is of potential utility,” says Richard Furie, MD, chief of the Division of Rheumatology at Northwell Health in Great Neck, New York. “Time will tell whether busy clinicians will utilize such a tool or resort to their current practice, which in my area is to refer patients based on the results of the ANA.”
Lara C. Pullen, PhD, is a medical writer based in the Chicago area.
References
- Adamichou C, Genitsaridi I, Nikolopoulos D, et al. Lupus or not? SLE Risk Probability Index (SLERPI): A simple, clinician-friendly machine learning-based model to assist the diagnosis of systemic lupus erythematosus. Ann Rheum Dis. 2021 Feb 10;annrheumdis-2020-219069. Online ahead of print.
- Aringer M, Costenbader K, Daikh D, et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019 Sep;71(9):1400–1412.