Kateryna Kon / shutterstock.com
The identification of specific transcription factors linked to the Epstein-Barr virus (EBV) provides new information on the molecular mechanisms underlying the link between EBV and autoimmune disease.
A recently published study, “Transcription Factors Operate Across Disease Loci, with EBNA2 Implicated in Autoimmunity,” co-led by John B. Harley, MD, PhD; Leah C. Kottyan, PhD; and Matthew T. Weirauch, PhD, at the Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Ohio, found EBNA2, a protein produced by Epstein-Barr, binds to several locations along the human genome—locations that are associated with seven autoimmune diseases: systemic lupus erythematosus (SLE), multiple sclerosis, rheumatoid arthritis (RA), juvenile idiopathic arthritis, inflammatory bowel disease, celiac disease and type 1 diabetes.1
Dr. Harley, director of the Center for Autoimmune Genomics and Etiology and professor of pediatrics and medicine at the University of Cincinnati, where he holds the David Glass Endowed Chair, says this finding is consistent with the possibility the Epstein-Barr virus causes these seven autoimmune diseases for at least some patients with these diseases.
“There were strong hints [of this] previously,” he says, “but the extent of the relationship was not known.”
“The link between EBV and autoimmunity has been suspected but remains controversial,” says Peter A. Nigrovic, MD, associate professor of medicine at Harvard Medical School, director of the Center for Adults with Pediatric Rheumatic Illness and director of the Joint Biology Consortium of Boston Children’s Hospital and Brigham and Women’s Hospital, Boston, commenting on the study. “This study integrates a vast amount of data to identify a genetic explanation for the connection. By providing a candidate mechanism, Harley and colleagues help render the epidemiologic evidence behind the association between EBV and autoimmunity more readily comprehensible.”
Uncovering Molecular Mechanisms
In the study, investigators developed and used two novel informatics tools. As explained in an article by Dr. Weirauch, a computational biologist, one of the tools is a new computational method for discovering disease-driving mechanisms.2 Called the Regulatory Element Locus Intersector, the method is an algorithm that estimates the significance of the intersection between disease-causing variants and DNA sequence. This tool allowed investigators to identify particular proteins occupying disease risk loci. Once these proteins were identified, the investigators used the second novel tool, the Measurement of Allelic Ratios Informatics Operator (MARIO), to provide insight into the molecular mechanisms of disease. Using MARIO, the investigators were able to show examples of a possible genetic role for the transcription factors linked to various diseases, Dr. Weirauch states in the article.
By using these tools, the investigators were able to show that transcription factors and cofactors are often concentrated in the risk loci of diseases and physiological phenotypes are frequently expected by chance, according to Dr. Harley.
Specifically, the study identified 2,264 relationships between hundreds of transcription factors and 94 phenotypes. For example, the study found the Epstein-Barr virus EBNA2 protein and many clustering human transcription factors occupy nearly half of systemic lupus erythematosus risk loci. The EBNA2 protein and clustering human transcription factors also occupy multiple other autoimmune diseases, including rheumatoid arthritis, multiple sclerosis, type 1 diabetes, juvenile idiopathic arthritis, inflammatory bowel disease and celiac disease. Overall, this association with EBNA2 and these autoimmune diseases demonstrates a gene-environment interaction of these diseases.
“The study by Dr. Harley and colleagues indicates the genes targeted by EBV are disproportionately ones that alter risk of autoimmune diseases, including lupus and rheumatoid arthritis,” says Dr. Nigrovic. “This finding, therefore, provides a new mechanistic link between EBV infection and autoimmune diseases.”
Some of the results for systemic lupus erythematosus from the recent study published in Nature Genetics are presented in Figure 1.
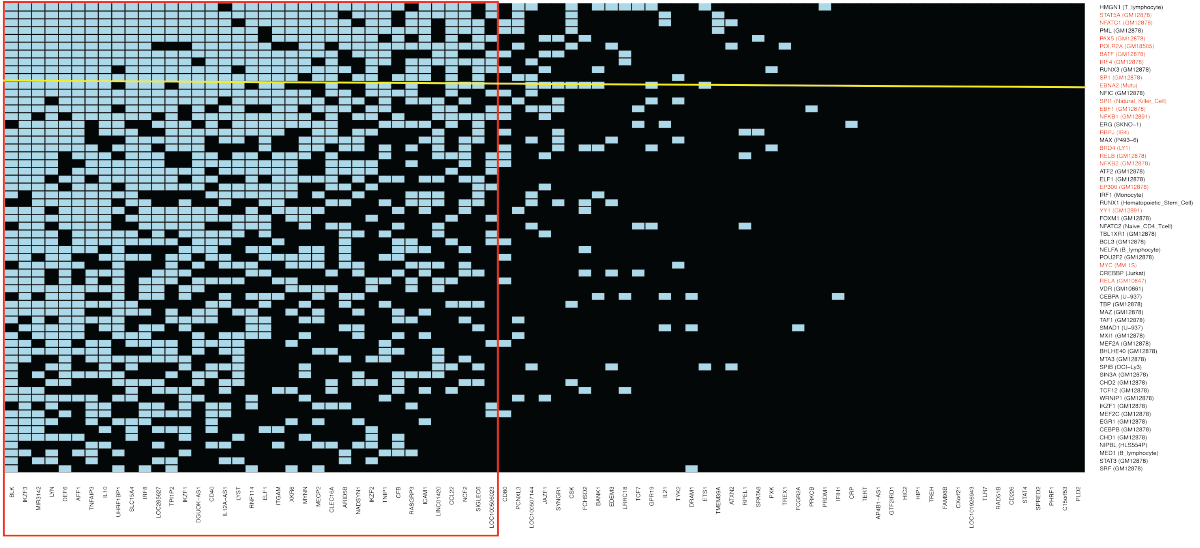
(click for larger image) Figure 1: Systemic Lupus Erythematosus
The blue boxes indicate the intersections found when transcription factors and co-factors (TFs) form complexes with variants located in 83 risk loci for systemic lupus erythematosus (SLE). The 83 SLE loci are from Europeans, African Americans, Asians and Hispanics. Each transcription factor is identified by its gene name; the cell type from which the DNA binding data were obtained are given in parentheses. Every TF shown has a probability of Pc<10-6 of the intersections with the SLE loci shown having occurred by chance after Bonferroni correction. The TFs in red are known to form super-enhancers in Epstein-Barr virus (EBV) transformed B cells. The EBV gene product EBNA2 is identified by a yellow line. All of the other TFs shown are encoded by the human genome. The red rectangle encloses a cluster of SLE loci and TFs that possibly function together to change the risk of SLE by gene expression regulatory mechanisms in B lymphocytes transformed by EBV. Note: Data are abstracted from Nature Genetics and adapted with permission by Sreeja Parameswaran.1
Clinical Implication?
This study points to the potential to develop future therapies that can manipulate the action of the proteins in persons harboring risk alleles at EBNA2-bound loci, conclude the study investigators.
Based on the specific findings of the study, the investigators plan to further investigate particular transcription factors and cell types for 94 phenotypes they’ve nominated to help provide a better understanding of the molecular and cellular origins of disease risk. “As new genetic association and TF [transcription factor] binding data are collected, approaches such as this will undoubtedly identify further disease mechanisms,” concludes Dr. Harley for the investigators.
The investigators are hoping experts on various diseases will share their results and collaborate to better understand these mechanisms. To that end, they have made their computer tools available online, along with their study data and results.
“My hope is that the study spurs further research into this connection,” says Dr. Nigrovic, “for example, to test experimentally the role of ENBA2 in the regulation of RA and lupus target genes.”
Most important to Dr. Nigrovic is what the study findings suggest may be the best target of all—prevention of autoimmunity. “To me, the most exciting implication is for prevention,” he says. “EBV infection increases with age, and ultimately, most people become infected. If we could prevent infection—by vaccination, for example—then it is possible we could reduce the incidence of autoimmune disease substantially. That’s an amazing prospect.”
Mary Beth Nierengarten is a freelance medical journalist based in Minneapolis.
References
- Harley JB, Chen X, Pujato M, et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet. 2018 May;50(5):699–707.
- Weirauch M. Expert offers inside look at new informatic tools developed for the Epstein-Barr virus study. Biomedical Informatics. 2018 Apr 16.