SAN DIEGO—John O’Shea, MD, chief of the molecular immunology and inflammation branch and scientific director at the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) in Bethesda, Md., provided an overview of the evolving understanding of the genome and epigenome that is opening new ways to look at the pathogenesis of rheumatic diseases and the potential for improved and better-targeted therapeutics in his talk at the 2013 ACR/ARHP Annual Meeting. In his presentation, “Deciphering Helper T Cell Identity: Finding Genomic Switches and Their Regulators,” he spoke about the new understanding of the genome that is now focused on learning why the small number of genes actually active in the genome get switched on or off and cause disease.
The Big Switch
The major premise of Dr. O’Shea’s talk was to describe the evolution in thought over an understanding of the genome, and how this evolution is changing an understanding of the genetic influence of diseases such as lupus and rheumatoid arthritis. The central dogma of molecular biology for years, he said, was that most of the genome was comprised of genes in which DNA makes RNA, which in turn makes proteins. We now know, he said, that in many cases the transcription stops at RNA so that much of the genome turns into RNA but doesn’t turn into protein.
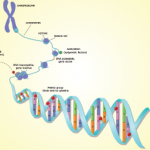
“The striking finding about a decade ago is that less than 2% of the genome is genes,” he said. What is now known is that, although only about 2% of the genome is genes, most of the genome is active and most of this activity appears to come from switches.
The big switch, Dr. O’Shea said, is that most of the genome appears to be switches that control this tiny portion of the genome that is genes. An important way that the switches seem to work is that the majority of the genome is translated into RNA, even though only a tiny portion of the genome is transcribed into RNA and translated in protein.
“The reason this is important is because, when we look at genetic associations with diseases like lupus and rheumatoid arthritis, often the parts of the genome associated with the risk for disease are not genes but, presumably, the switches,” he said, adding that, in general, “the risk of autoimmune disease most often maps to places in the genome that does not code for typical protein-coding genes.”
Given this new understanding, he said the challenge is to understand where all the switches are, what they regulate, and how they are regulated.
More Refined View of the Genome
Dr. O’Shea opened his talk with a metaphor to illustrate how the focus on switches versus genes has refined the current understanding of the genome and its association with diseases. He said the old way of looking at the genome was like a Neanderthal man coming into the lecture hall who, when asked to turn off the lights, would try to climb up to the ceiling to smash the light itself, not knowing that there are switches on the wall for turning off the light. The current view that switches control gene expression now focuses investigation on finding the switches. What makes this even more interesting and challenging is that even if a switch is near a gene, it may or may not regulate that gene. The switch that regulates a gene may be near or far from the gene.
Important types of switches in the genome that regulate genes from a distance are known as enhancers, and a subset of enhancers known as super-enhancers contain huge numbers of regulatory elements. According to Dr. O’Shea, the genetic risk of various types of disease appears to be enriched in these super-enhancer areas.
“All of us have switches, but a sequence of your switches and my switches may not be the same, and that may affect how genes are regulated,” he said, adding that this may determine the different genetic risk of disease among people.
Added to the complexity of disease development, he said, is the role played by the environment. “The other part of the epigenome is that it is responsive to the environment, so the epigenome can be modified,” he said. “It is not just the genes, it is how the genes are allowed to be turned on, silenced, or in a configuration that allows them to be active.”
Learning From Oncology
Although applying this view of the genome to a better understanding of the pathogenesis of rheumatic diseases and finding new therapeutic agents for these diseases may be relatively new to rheumatologists, Dr. O’Shea said that oncologists have already developed drugs based on their efforts to understand the epigenome in cancer.
Noting that a number of original drugs currently used to treat rheumatic diseases, such as methotrexate, were first used to treat cancer, Dr. O’Shea said, “This may be another circumstance where rheumatologists learn from oncologists.”
Mary Beth Nierengarten is a freelance medical journalist based in St. Paul, Minn.