Oxidative Stress
Mitochondrial dysfunction has also been described in SLE T cells. This dysfunction leads to the increased production of destructive reactive-oxygen intermediates and oxidative stress. The mechanisms and consequences of this abnormality are complex and not fully understood. SLE T cells show increased activity of mammalian target of rapamycin (mTOR), a kinase that regulates mitochondrial transmembrane potential. Among other effects, this increased activity results in abnormal calcium flux and contributes to decreased CD3ζ expression, the central signaling defect in SLE T cells. Treating SLE T cells with rapamycin, an mTOR inhibitor, restores CD3ζ expression and normalizes downstream signaling.19 Rapamycin also improves the disease phenotype and prolongs survival of lupus-prone mice. In humans, rapamycin treatment showed some benefit in the treatment of nine SLE patients with refractory disease, but more investigation will be needed to determine whether this will be useful for patients more broadly.20
Future Directions
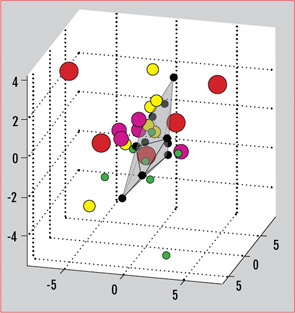
The myriad T-cell abnormalities that have been described with SLE make for a confusing, complicated picture of immune dysregulation. When considered individually and in vitro, many of these abnormalities vary with disease activity or recapitulate certain aspects of the disease in vivo. The SLE patient population in practice, however, is a clinically and genetically heterogeneous group. There is no one single marker that is useful practically as a biomarker for this diverse population. Recently, our group has described the use of a gene expression array that can capture a broader picture of T-cell dysfunction. This is a single test that allows the simultaneous measurement of expression levels for a panel of genes. We selected 30 genes previously reported to have aberrant expression or function in SLE, including CREM, Syk, CD3ζ, and other factors mentioned in this review. Analysis of gene-expression patterns showed that the SLE samples segregated from normal controls.21 Differential patterns of expression also emerged that were specific for the type of organ system involvement. As therapies targeting various aspects of T-cell dysfunction come into clinical use, these gene expression patterns could also be used to predict responses to specific treatments. (See Figure 3)
New research into epigenetic regulation of gene expression also may provide further insight into how environmental factors may come into play in the pathogenesis of autoimmunity. Several studies of DNA methylation in T cells have shown significant differences in methylation patterns between SLE T cells compared with normal controls.22,23 In general, SLE T cells show lower methylation levels for many genes, including some previously implicated in SLE pathogenesis. As an example, our group has shown that increased expression of protein phosphatase 2A (PP2A, an enzyme thought to contribute to the decreased IL-2 phenotype in SLE), is due to hypomethylation of its promoter.24 Less is known about histone modification in autoimmunity, although this is also an area of active research. Histone deacetylase inhibitors are already in development for treatment of certain cancers, and there are promising studies of these compounds in mouse models of lupus.25