ellepigrafica / shutterstock.com
CHICAGO—Because the epigenome has been implicated in a variety of rheumatic conditions, a Basic Research Conference was convened on Epigenetics in Immune-Mediated Disease in conjunction with the 2018 ACR/ARHP Annual Meeting.
Melanie Ehrlich, PhD, professor of human genetics and genomics at Tulane University School of Medicine, New Orleans, opened the conference. She has a long association with epigenetics, having founded the DNA Methylation Society (now the Epigenetics Society) in 1994.
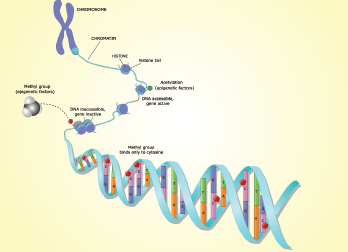
Dr. Ehrlich introduced the topic of epigenetics by explaining that although all cells in an organism have the same DNA sequence, different types of cells have different remembered epigenetic profiles layered on top of their genetic profiles. These cell type-specific epigenetic profiles allow cells to have long-lasting differentiation-dependent differences in gene expression. Epigenetics is thus the inheritance from cell to cell of these DNA modification or chromatin modification profiles. The epigenetic profiles are established via changes in chromatin or DNA, but not changes in the actual DNA sequence (A, T, C or G).
Dr. Ehrlich described two main forms of vertebrate epigenetics: inherited DNA modifications and inherited chromatin structure. The DNA modifications in mammals are primarily DNA methylation or, less frequently, hydroxymethylation of C residues. In contrast, chromatin modifications are chemical alterations of histones or stable replacements in chromatin of certain histones, non-histone proteins or non-coding RNAs. All of the epigenetic changes occur only at certain DNA sequences in the genome and are often cell-type specific.
Alterations in DNA methylation play important roles not only in cellular differentiation, Dr. Ehrlich explained, but also in disease. She urged the gathered rheumatologists to remember many differentiation- or disease-related DNA methylation changes occur at little-studied enhancers rather than at the more frequently examined gene promoter. Moreover, not all DNA methylation changes have a biological effect. Many epigenetic changes are bystander changes and must be viewed in a larger context. She left the audience with parting words of advice, “The devil is in the details!”
Epigenetics & the Immune System
John J. O’Shea, MD, scientific director at the National Institutes of Health’s National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, Md., described the intersection of epigenomics and lymphocytes. He explained how enhancers, super-enhancers and chromatin accessibility contribute to lymphocyte development and activation. Thus, they can affect disease and the response to drugs used to treat disease. He reiterated the message, “The complexity of organisms relates, in a way, better to junk DNA,” adding, “There is a lot of transcription going on in the genome that extends way beyond the few canonical genes that we have.”
Dr. O’Shea also explained that research has revealed terminal differentiation is no longer terminal, as evidenced by inducible pluripotent stem cells. Moreover, as technology has exploded with massive parallel sequencing, it has revealed many new marks on DNA, some of which are relatively stable and many of which are not. He described epigenetics as adaptations of chromosomal regions that perpetuate altered activity. These epigenetic mechanisms have been defined as transducing the inheritance of gene expression and, therefore, are critical for understanding the behavior of immune cells.
His team became enthralled with the idea the genome consists of thousands of tissue-specific switches. “We were sort of fascinated by this,” he explained. “How might this concept change our views of lymphocyte activation and the action of cytokines?” Their research revealed about 20,000 switches in naive T cells, and approximately 90 switches are shared among lymphocytes, macrophages and embryonic stem cells. When Dr. O’Shea and his colleagues looked more closely at these switches, they realized the distinct fates of T cells represent an integration of environmental and cytokine signals through signal-dependent transcription factors (SDTFs) and the action of intrinsic lineage-defining transcription factors (LDTFs). Signal transducer and activator of transcription 1 (STAT1) is an example of such an SDTF. It is directly activated by interferon gamma (IFNɣ) and induces transactivation of its targets.

