ellepigrafica / shutterstock.com
CHICAGO—Because the epigenome has been implicated in a variety of rheumatic conditions, a Basic Research Conference was convened on Epigenetics in Immune-Mediated Disease in conjunction with the 2018 ACR/ARHP Annual Meeting.
Melanie Ehrlich, PhD, professor of human genetics and genomics at Tulane University School of Medicine, New Orleans, opened the conference. She has a long association with epigenetics, having founded the DNA Methylation Society (now the Epigenetics Society) in 1994.
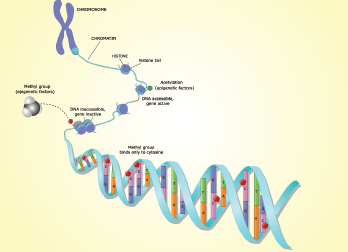
Dr. Ehrlich introduced the topic of epigenetics by explaining that although all cells in an organism have the same DNA sequence, different types of cells have different remembered epigenetic profiles layered on top of their genetic profiles. These cell type-specific epigenetic profiles allow cells to have long-lasting differentiation-dependent differences in gene expression. Epigenetics is thus the inheritance from cell to cell of these DNA modification or chromatin modification profiles. The epigenetic profiles are established via changes in chromatin or DNA, but not changes in the actual DNA sequence (A, T, C or G).
Dr. Ehrlich described two main forms of vertebrate epigenetics: inherited DNA modifications and inherited chromatin structure. The DNA modifications in mammals are primarily DNA methylation or, less frequently, hydroxymethylation of C residues. In contrast, chromatin modifications are chemical alterations of histones or stable replacements in chromatin of certain histones, non-histone proteins or non-coding RNAs. All of the epigenetic changes occur only at certain DNA sequences in the genome and are often cell-type specific.
Alterations in DNA methylation play important roles not only in cellular differentiation, Dr. Ehrlich explained, but also in disease. She urged the gathered rheumatologists to remember many differentiation- or disease-related DNA methylation changes occur at little-studied enhancers rather than at the more frequently examined gene promoter. Moreover, not all DNA methylation changes have a biological effect. Many epigenetic changes are bystander changes and must be viewed in a larger context. She left the audience with parting words of advice, “The devil is in the details!”
Epigenetics & the Immune System
John J. O’Shea, MD, scientific director at the National Institutes of Health’s National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, Md., described the intersection of epigenomics and lymphocytes. He explained how enhancers, super-enhancers and chromatin accessibility contribute to lymphocyte development and activation. Thus, they can affect disease and the response to drugs used to treat disease. He reiterated the message, “The complexity of organisms relates, in a way, better to junk DNA,” adding, “There is a lot of transcription going on in the genome that extends way beyond the few canonical genes that we have.”
Dr. O’Shea also explained that research has revealed terminal differentiation is no longer terminal, as evidenced by inducible pluripotent stem cells. Moreover, as technology has exploded with massive parallel sequencing, it has revealed many new marks on DNA, some of which are relatively stable and many of which are not. He described epigenetics as adaptations of chromosomal regions that perpetuate altered activity. These epigenetic mechanisms have been defined as transducing the inheritance of gene expression and, therefore, are critical for understanding the behavior of immune cells.
His team became enthralled with the idea the genome consists of thousands of tissue-specific switches. “We were sort of fascinated by this,” he explained. “How might this concept change our views of lymphocyte activation and the action of cytokines?” Their research revealed about 20,000 switches in naive T cells, and approximately 90 switches are shared among lymphocytes, macrophages and embryonic stem cells. When Dr. O’Shea and his colleagues looked more closely at these switches, they realized the distinct fates of T cells represent an integration of environmental and cytokine signals through signal-dependent transcription factors (SDTFs) and the action of intrinsic lineage-defining transcription factors (LDTFs). Signal transducer and activator of transcription 1 (STAT1) is an example of such an SDTF. It is directly activated by interferon gamma (IFNɣ) and induces transactivation of its targets.
Alterations in DNA methylation play important roles not only in cellular differentiation, but also in disease.
SDTFs and LDTFs thus bind to enhancers distributed asymmetrically throughout the genome. Genomic regions of dense deposition are referred to as super-enhancers, and these super-enhancers are linked to cell identity. For example, in many cases, the loci identified via genome-wide association studies hits are enriched for super-enhancer architecture, and Dr. O’Shea showed data indicating super-enhancers reflect both cell identity and cell state.
Rheumatologists should be aware of these super-enhancers because “the drugs that we are giving patients are more likely to affect super-enhancer regions,” noted Dr. O’Shea. “Deciphering all these switches in immune cells is an enormous challenge, but in the future, better knowledge of these numerous regulatory elements should create tremendous opportunities for new treatments.” For example, Dr. O’Shea and colleagues have found drugs like tofacitinib preferentially affect rheumatoid arthritis (RA) risk genes with super-enhancer structure, as opposed to those with typical enhancer architecture.
Esteban Ballestar, PhD, senior group leader of the Cancer Epigenetics and Biology Program at the Bellvitge Biomedical Research Institute, Barcelona, expanded the conversation to B cells and antibodies. In particular, he described the DNA methyltransferase family of enzymes, which include ten-eleven translocation (TET) enzymes that actively demethylate DNA. Dr. Ballestar reiterated DNA methylation is linked to gene expression, and he specifically discussed common variable immunodeficiency, which is the most frequent symptomatic primary antibody deficiency. It has an incidence of 1 in 25,000 and is caused by a severe deficiency of switched memory B cells and a marked decrease in IgG. Closer examination has revealed “these patients not only have a smaller number of these memory cells, but they have a defect in demethylating these genes,” explained Dr. Ballestar.
Role of Aging in Autoimmunity
Raymond Yung, MD, director of the Geriatrics Center and Institute of Gerontology at the University of Michigan, Ann Arbor, spoke about the role of aging in autoimmunity. Dr. Yung is also the Jeffrey B. Halter Professor of Geriatric Medicine, chief of the Division of Geriatric and Palliative Medicine, director of the University of Michigan Institute of Gerontology and Geriatrics Center at Michigan Medicine. Although aging is associated with an increased prevalence of serological autoimmunity, the increase, surprisingly, does not necessarily translate into a high incidence of full-blown autoimmune disease in old age. Dr. Yung examined the paradox and found evidence that suggested epigenetics is an important process that influences aging and may serve as the link between aging and chronic diseases.
Dr. Yung used the term inflamm-aging to describe how, as humans age, blood and serum have increased levels of interleukin 6 (IL-6), IL-8, tumor necrosis factor (TNF) and C-reactive protein. Although it is not yet obvious why these cytokines increase as people age, Dr. Yung put forth the hypothesis the body encounters numerous infectious agents and develops multiple chronic infections as it ages, and these infections stimulate the immune system. “Over time, the body reacts by not returning to baseline,” suggested Dr. Yung. He noted other factors beyond infection could also be driving this phenomenon, including obesity, microbiota, renin-angiotensin, hormonal changes, redox stress, telomere dysfunction and glycation.
Inflamm-aging extends beyond a shift in naive and memory subsets to include an enlargement of fat-associated lymphoid clusters, as well as an increase in adipose tissue macrophages. Dr. Yung concluded, “The immune system is complicated. Aging is complicated. When you put the two of them together, it gets really complicated.”
Epigenetics Meets Rheumatology
Nan Shen, MD, PhD, director of Shanghai Institute of Rheumatology, took the stage to speak about non-coding genes that account for most of the transcription of the genome. He described these non-coding sequences as quite complex and walked the audience through a (relatively) simple description. He explained the long non-coding RNAs (lncRNAs) in the nucleus and cytoplasm have multiple functions, including epigenetic modification, transcriptional regulation, post-transcriptional regulation and post-translational regulation. They thus regulate gene expression at multiple levels by interacting with DNA, RNA and protein. Perhaps of most interest to rheumatologists, Dr. Shen stated, they are also critical for immune cell development, activation and function. They are likely to be especially important in rheumatology because several lncRNAs are dysregulated in rheumatic diseases.
Dr. Shen and others have focused their efforts on identifying the lncRNAs relevant to systemic lupus erythematosus (SLE). They have used several strategies, including data mining, to identify the lncRNAs involved in immune system regulation, those associated with disease activity, those differentially expressed between patients with lupus and healthy controls, and those located near genetic susceptibility loci. Their efforts have yielded an lncRNA stimulated by interferon that appears to be important in the pathophysiology of SLE.
Epigenetics & Psoriasis
Qianjin Lu, MD, director of the Institute of Dermatology at Central South University, Changsha, Hunan, China, discussed the role of epigenetics in a mouse model of psoriasis. He noted the literature has documented monozygotic twins who are discordant for psoriasis.
Psoriasis is associated with inflammatory cell infiltration, increased cytokine production and hyperproliferation of keratinocytes. Both genetic and environmental factors create the epigenetic profile of psoriasis, demonstrating that DNA methylation changes gene expression. Moreover, skin biopsies of patients with psoriasis reveal higher global DNA methylation, and researchers have now been able to use epigenome-wide association analysis to identify nine skin DNA methylation loci for psoriasis. In addition, high throughput data analysis has identified methylated genes from different clinical specimens of psoriasis patients. More detailed studies have revealed the DNA in skin lesions and peripheral blood mononuclear cells (PBMCs) of patients with psoriasis are hypermethylated and include abnormal histone modifications.
Non-coding microRNA (miRNA) also plays a role in the modulation of immunological mechanism of psoriasis. Dr. Lu’s group has found several key miRNAs, such as miRNA-210, are specifically overexpressed in psoriasis, whereas other known miRNAs are specifically downregulated in psoriasis. A closer examination of immune cells revealed miRNA-210 expression is elevated in CD4+ T cells, as well as in skin lesions from patients with psoriasis. Moreover, expression levels of miRNA-210 correlate with disease severity. The research by Dr. Lu and colleagues suggests miRNA-210 contributes to the altered balance between pathogenic Th1/Th17 cells and Th2 cells in patients with psoriasis and accelerates the development of disease. Their results thus detail a crucial role for miRNA-210 in the immune imbalance of T lymphocyte subsets in psoriasis and suggest a potential therapeutic approach.
From Bench to Bedside
The session concluded with presentations on two abstracts from the conference. Chris M. Dunn, a master’s student at the University of Oklahoma Health Sciences Center, Oklahoma City, presented his research, titled, “Epigenetic Editing of FOXP3 in Human T Cells Is Sufficient to Induce Overexpression and Create a Regulatory T Cell Phenotype in Vitro.”
Mr. Dunn described the epigenetic editing of the transcription factor FOXP3 using a dCas9-TET1 construct. He acknowledged, “There is a hot debate on whether FOXP3 expression and CTLA4 expression are related in any way.” Nevertheless, his team was able to induce DNA demethylation, overexpression and a regulatory T cell phenotype.
“Our data are intriguing but need confirmation, particularly to clarify the persistence of induced DNA methylation changes and resistance to phenotype switching. If confirmed, this approach has the potential to significantly improve upon current methods of Treg [regulatory T cell] generation,” he explained.
Jieun Shin, PhD, with the Research Institute of Chong Kun Dang Pharmaceutical Corp., Yongin, Korea, presented her group’s research on CKD-506, a first-in-class selective and potent histone deacetylase (HDAC) 6 inhibitor. Dr. Shin reported data from ex vivo studies of RA patient samples, as well as data from the rat adjuvant-induced arthritis (AIA) model. She began her presentation by describing a macrophage cell line that overexpressed HDAC6 and had increased production of inflammatory cytokines. When she and her colleagues treated these cells with CKD-506, they were able to inhibit the inflammatory mediators and inhibit NF-kB and AP-1 signaling in the cells.
When the investigators treated RA patient samples with CKD-506, they were also able to inhibit inflammatory mediators, specifically TNF-α. In addition, CKD-506 induced IL-10 production when RA patient peripheral blood mononuclear cells were stimulated with lipopolysaccharide. When they looked at cellular function, they found CKD-506 induced CTLA4 and enhanced Treg activity in PBMCs from patients with RA. Specifically, CD4+CD25– cells differentiated into Treg cells in the presence of CKD-506. Dr. Shin and her colleagues then orally administered CKD-506 to AIA rats and found it not only repressed arthritis in the rats, but it also acted synergistically with methotrexate.
CKD-506 improved symptoms of RA via regulation of inflammation and T cell function, according to Dr. Shin. CKD-506 is currently in preparation for a phase 2a clinical trial in the European Union for the treatment of patients with moderate to severe RA. The team is also exploring the possibility CKD-506 will be effective in the treatment of inflammatory bowel disease.
Something to Think About
The five-hour session led the rheumatologists on a journey from epigenetics in physiology to epigenetics in pathophysiology to a description of a drug with an epigenetic target poised to enter a phase 2 trial. The journey was exhausting, and many members of the audience were also battling jet lag, but they left the session with the air of visitors who had been exposed to a newly discovered wing of the castle. There was excitement and the sense the session gave them a lot to think about.
Lara C. Pullen, PhD, is a medical writer based in the Chicago area.

