Struggling to convince the Royal Society, van Leeuwenhoek observed the plaque between his own teeth “with great wonder, that in the said matter there were many very little living animalcules, very prettily a-moving.”
It would take two centuries, however, to prove these animalcules responsible for the origin of infectious processes. With Koch and Pasteur leading and rationalizing the nascent field of microbiology, most medical disciplines then turned into an “infectious rush era.” The logic followed that, since bacteria caused all cardinal signs of inflammation in affected sites, rheumatic diseases were also inevitably a deleterious effect of microorganisms. Theories flourished, and multiple lines of investigation suggested a link between oral or intestinal microbes and RA. Soon after, arthritis deformans was considered to be due to tuberculosis; the focal sepsis hypothesis claimed that RA was caused by periodontal flora; and the toxemic factor idea proposed a role in diseases of substances produced by intestinal bugs.1,7-9 The first half of the 20th century, without the means to prove any of these hypotheses (mainly because one in five bacteria are noncultivable), saw thousands of teeth extractions and even dozens of colectomies performed, hoping to achieve the ultimate cure for RA. Ultimately, none of those expectations materialized, and surgical solutions for RA were gradually abandoned.
Most recently (although debated), paleopathological and epidemiologic evidence has suggested that RA originated in the New World and spread through a transmissible vector to the rest of the globe after 1492.10 These conclusions are based on the lack of skeletal remains with RA-like features outside of America and the high incidence of disease in Amerindians compared with Caucasians, Europeans, Africans, and Asians. Clinical studies implicating specific bacterial agents as triggers for RA have mostly relied on indirect evidence (serological and biased polymerase chain reaction methods) and circumstantial observations.
Who Needs Culture, Anyway?
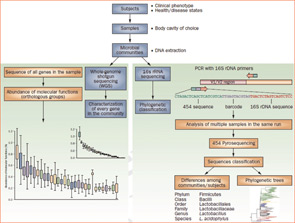
What theoretically appeared reasonable and necessary to advance the understanding of the microbiome has, nevertheless, encountered challenges. Central to these challenges is the inability to culture and grow approximately 80% of bacteria found in our microbiomes. Whether related to oxygen conditions or nutritional requirements, the immense majority of bacteria cannot survive if they leave their natural habitats. Because this deportation means death for many species, the DNA sequencing technology revolution has enabled the unimaginable: to bypass the need to isolate living organisms for their identification (see Figure 2).
Figure 2
Culture-independent genomic analysis of the human microbiome. Culture-independent techniques have advanced our capacity to survey complex microbial communities in human samples. Well-characterized individuals (healthy and diseased) are asked to donate samples for microbiome analyses. Two metagenomic sequencing approaches are utilized. Conserved and variable 16S rRNA genomic regions are amplified and subjected to pyrosequencing. The resulting sequences are then aligned, filtered, and compared with publicly available databases of 16S rRNA sequences, enabling taxonomic classification of bacteria present or absent in a given sample. Whole genome shotgun sequencing provides information that enables identification of genes present and allows for subsequent comparison of enzymatic pathways and functions represented among different samples. Enzymatic databases are also available to assist in the identification of protein function, enabling the richness and diversity of functional capacities provided by the microbiome to be assessed.